In this section, we’ll utilize knowledge we learnt from the last section (see here), to implement a ResNet Network (paper).
Note that we follow the original paper’s work. Our implementation is a simper version of the official torchvision implementation. (That is, we only implement the key structure, and the random weight init. We don’t consider dilation or other things).
Preliminaries: Calculate the feature map size
- Basic formula
Given a convolution kernel with size K, and the padding P, the stride S, feature map size I, we can calculate the output size as O = ( I - K + 2P ) / S + 1.
- Corollary
Based on the formula above, we know that when S=1:
- K=3, P=1 makes the input size and output size same.
- K=1, P=0 makes the input size and output size same.
Overall Structure
The Table 1 in the original paper illustrates the overall structure of the ResNet:
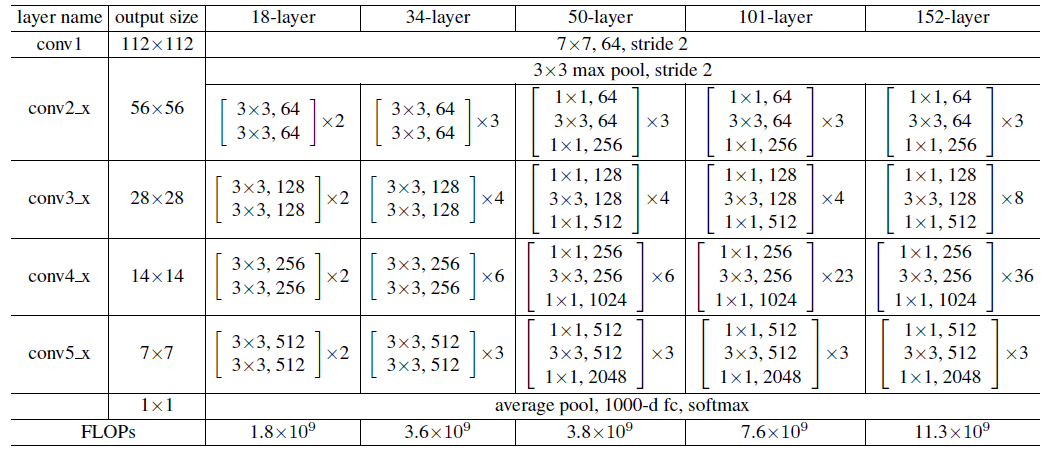
We know that from conv2, each layer consists of many blocks. And the blocks in 18, 34 layers is different from blocks in 50, 101, 152 layers.
We have several deductions:
- When the feature map enters the next layer, the first block need to do a down sampling operation. This is done by setting the one of the convolution kernel’s
stride=2. - At other convolution kernels, the feature map’s size is same. So the convolution settings is same as the one referred in Preliminaries.
Basic Block Implementation
The basic block’s structure looks like this:
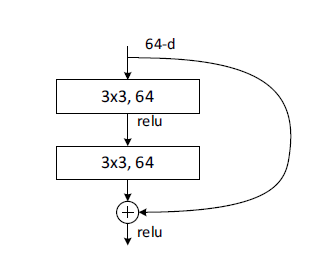
Please see the code below. Here, apart from channels defining the channels in the block, we have three additional parameters, in_channels, stride, and downsample to make this block versatile in the FIRST block in each layer.
According to the ResNet structure, for example, the first block in layer3 has the input 64*56*56. The first block in layer3 has two tasks:
- Make the feature map size to
28*28. Thus we need to set its stride to2. - Make the number of channels from
64to128. Thus thein_channelshould be64. - In addition, since the input is
64*56*56, while the output is128*28*28, we need a down sample convolution to match the shortcut input to the output size.
1 | import torch |
Bottleneck Block Implementation
The bottleneck block’s structure looks like this:
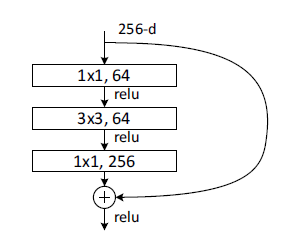
To reduce the computation cost, the Bottleneck block use 1x1 kernel to map the high number of channels (e.g., 256) to a low one (e.g., 64), and do the 3x3 convolution. Then, it maps the 64 channels to 256 again.
Please see the code below. Same as the basic block, We have three additional parameters, in_channels, stride, and downsample to make this block versatile in the FIRST block in each layer. The reasons are same as above.
1 | class ResidualBottleNeck(nn.Module): |
ResNet Base Implementation
Then we can put thing together to form the ResNet model! The whole structure is straight-forward. We define the submodules one by one, and implement the forward() function.
There is only two tricky point:
- To support the
ResNetBasefor two different base blocks, the base block can be passed to this initializer. Since two base blocks have slightly differences in setting the channels,ResidualBasicBlockandResidualBottleNeckhave an attribute calledexpansion, which convenient the procedure in setting the correct number of channels and outputs. - See the
_make_layerfunction below. It need to determine whether we need to do the down sample. And the condition and explanation is described below.
1 | class ResNetBase(nn.Module): |
Encapsulate the Constructors
Finally, we can encapsulate the constructors by functions:
1 | def my_resnet18(in_channels=3): |
Then, we can use it as normal models:
1 | img = torch.randn(1, 3, 224, 224) |